Introductions Introductions
|
|
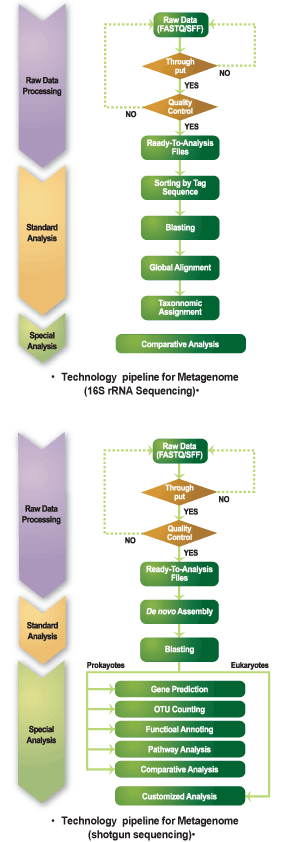
Metagenomics is the study of the variation of species in a complex microbial sample. Using next generation sequencing it is possible to profile and investigate microbial communities by sequencing the marker gene of choice (for example, 16S rRNA) or the whole microbial genome.
|
|
|
MACROGEN provides integrated microbial NGS services with high-quality data and rapid turnaround time. The microbial-specialized system provides efficient sequencing workflow from various sample sources on different sequencing platforms. The analysis of bioinformatics data includes detailed gene assembly, gene annotation, and taxonomic assignment of the identified microbiome.
|
|
|
|
16S(18S) rRNA Sequencing
|
|
Metagenomic 16S (or 18S) rRNA sequencing enables efficient identification of bacterial and archaeal microorganism diversity in a specific environment.
|
|
|
Whole Genome Shotgun Sequencing
|
|
Metagenomic shotgun sequencing is a comprehensive sampling method of all the genes in all the organisms present in a given mixed sample. The method enables to evaluate microbial diversity and detect the abundance of species under different conditions. For shotgun sequencing, DNA is purified and randomly sheared into smaller fragments before sequencing.
|
|
|
|
 Sequencing platform Sequencing platform
|
• HiSeq2000/2500
• MiSeq system
• Ion PGM
|
|
|
 Data Analysis* Data Analysis*
|
|
16R (or 18S) rRNA Sequencing
|
• Global Alignment
• Taxonomic Assignment
• OTU Counting
• Comparative Analysis
|
|
|
|
Whole Genome Shotgun Sequencing
|
• De novo Assembly
• BlastX
• Gene Prediction
• Functional Assignment
• OTU Counting
• Statistical Analysis
|
|
|
|
*Data Analysis may vary depending on the avaliability of reference and the type of platform.
|
|
For any inquiry of NGS service, please feel free to contact us.
E-mail : ngs.apac@macrogen.com
|




 Privacy policy
Privacy policy Terms & Conditions
Terms & Conditions Introduction
Introduction Sample Submission Guide
Sample Submission Guide Introduction
Introduction Oligo Delivery Tip
Oligo Delivery Tip Oligo DNA design tool
Oligo DNA design tool


 Introductions
Introductions